Abstract
Introduction
Patients (pts) with relapsed or refractory large B-cell lymphomas (rrLBCL) can achieve long-term remission after CD19 chimeric antigen receptor T-cell therapy (CART), but more than half of recipients will experience treatment failure. High pretreatment tumor burden by elevated LDH, metabolic tumor volume, or circulating tumor DNA (ctDNA) levels by clonotype sequencing are associated with inferior outcomes. Low pass whole genome sequencing (lpWGS) is a highly simplified assay capable of identifying copy number alterations (CNA) from cell free DNA (cfDNA) in blood and requires less time, cost, and sample volume to perform vs. other cfDNA sequencing methods; it may be an efficient and precise proxy for tumor burden to identify pts at high risk of progression after CART. Here, we performed lpWGS on pretreatment plasma samples from pts who received CART for rrLBCL and incorporated it into a prognostic risk model.
Methods
Pts with rrLBCL treated with standard-of-care CART between 5/2018 and 2/2021 at MDACC with plasma samples from time of apheresis were retrospectively identified. cfDNA was extracted from plasma, DNA libraries constructed, and lpWGS performed with a mean coverage of 1.3X. CNA segmentation was generated by CopyWriteR and CNApp (Franch-Expósito et al. Elife 2020) was used to compute scores for focal (FCS, <50% chromosome arm affected), broad (BCS, ≥50%), and relative global (GCS) CNA burdens. Baseline pt characteristics were recorded at time of apheresis. Co-primary endpoints were progression free survival (PFS) from CART infusion and rate of durable response, defined as achieving a complete or partial response without a PFS event with ≥ 3-month follow up. A regularized cox regression model and recursive partitioning analysis tested all covariates against PFS and durable response rate, respectively.
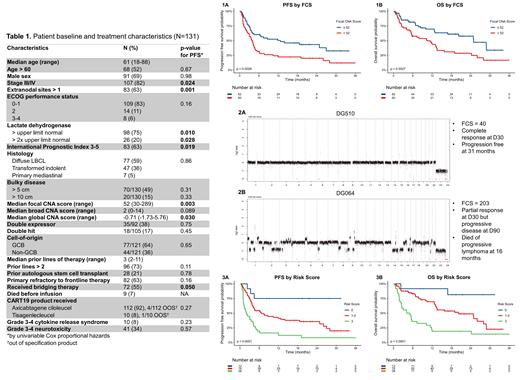
Results
A total of 135 pts had available plasma collected from the apheresis timepoint, of which 131 (97%) were successfully sequenced and included in the analysis. A total of 122 (93%) pts received their CART product and 9 (7%) died before infusion. Median follow up was 20.7 mo (range 0.5-36) with data cutoff 6/15/2021. Pt characteristics at time of apheresis are described in Table 1. A total of 83 (68%) treated pts progressed and 68 (56%) died. Median PFS and OS were 4.5 and 13.5 mo, respectively, with 1-year PFS and OS rates of 36% and 57%. ECOG PS > 1 was the only covariate associated with death before infusion (p<0.0001). A total of 38 of 121 (31%) evaluable pts (excluding 9 who died before CART and 1 lost to follow up early) were durable responders. Associations between covariates and PFS by univariable analysis are shown in Table 1. Multivariable selection for PFS determined that the 3 independent covariates that comprised the best model were > 1 extranodal (EN) site of disease, LDH > upper limit normal (ULN), and FCS > 52 (median). FCS > 52 was associated with inferior PFS (p=0.0029, HR 1.93 95% CI 1.24-2.99, Fig. 1A), durable response rate (44 v 18%, p=0.0031), and OS (p=0.0027, HR 2.09 95% CI 1.28-3.41, Fig. 1B). Representative genome-wide CNA profiles of 2 pts with FCS of 40 (Fig. 2A) and 203 (Fig. 2B) are included. We created a risk score from 0-3 where pts received 1 point for each negative prognostic feature met (> 1 EN site, LDH > ULN, and FCS > 52), then stratified pts by low (0), intermediate (1-2), or high risk (3). PFS (p<0.0001, Fig. 3A), durable response rate (75 v 32 v 10%, p<0.0001) and OS (p<0.0001, Fig. 3B) were significantly different when comparing low, intermediate, and high-risk pts.
Discussion
This retrospective analysis is the first to incorporate lpWGS of pretreatment cfDNA to determine CNA burden and associate it with outcomes after CART therapy. The FCS score is associated with survival and is likely a surrogate for ctDNA and tumor burden due to variability in tumor purity. Having > 1 EN site of disease and LDH > ULN were also independently associated. A simple risk model using the 3 variables can reliably risk stratify CART pts prior to therapy, should be validated in an independent/prospective dataset, and could achieve the goal of identifying high-risk pts for interventional studies. lpWGS may therefore represent a more time-, cost-, and sample-efficient method of utilizing pt plasma vs. alternative sequencing methods.
Fowler: BostonGene, Corp: Current Employment, Current holder of stock options in a privately-held company; Bristol Myers Squibb, F. Hoffmann-La Roche Ltd, TG Therapeutics and Novartis: Membership on an entity's Board of Directors or advisory committees, Research Funding. Parmar: Cellenkos Inc.: Current holder of individual stocks in a privately-held company, Membership on an entity's Board of Directors or advisory committees, Patents & Royalties, Research Funding. Ahmed: Merck: Research Funding; Seagen: Research Funding; Xencor: Research Funding; Tessa Therapeutics: Membership on an entity's Board of Directors or advisory committees, Research Funding. Steiner: Seattle Genetics: Research Funding; Rafael Pharmaceuticals: Research Funding; BMS: Research Funding. Samaniego: Imbrium: Membership on an entity's Board of Directors or advisory committees; Arog: Research Funding. Nastoupil: MorphoSys: Honoraria; Pfizer: Honoraria, Research Funding; Takeda: Honoraria, Other: DSMC, Research Funding; Janssen: Honoraria, Research Funding; Denovo Pharma: Other: DSMC; ADC Therapeutics: Honoraria; TG Therapeutics: Honoraria, Research Funding; Genentech: Honoraria, Research Funding; IGM Biosciences: Research Funding; Caribou Biosciences: Research Funding; Novartis: Honoraria, Research Funding; Bayer: Honoraria; Epizyme: Honoraria, Research Funding; Bristol Myers Squibb/Celgene: Honoraria, Research Funding; Gilead/Kite: Honoraria, Research Funding. Neelapu: Kite, a Gilead Company, Bristol Myers Squibb, Merck, Poseida, Cellectis, Celgene, Karus Therapeutics, Unum Therapeutics (Cogent Biosciences), Allogene, Precision BioSciences, Acerta and Adicet Bio: Research Funding; Kite, a Gilead Company, Merck, Bristol Myers Squibb, Novartis, Celgene, Pfizer, Allogene, Kuur, Incyte, Precision BioSciences, Legend, Adicet Bio, Calibr, and Unum Therapeutics: Other: personal fees; Kite, a Gilead Company, Merck, Bristol Myers Squibb, Novartis, Celgene, Pfizer, Allogene Therapeutics, Cell Medica/Kuur, Incyte, Precision Biosciences, Legend Biotech, Adicet Bio, Calibr, Unum Therapeutics and Bluebird Bio: Honoraria; Takeda Pharmaceuticals and related to cell therapy: Patents & Royalties. Strati: Astrazeneca-Acerta: Research Funding; Roche-Genentech: Consultancy. Westin: Curis: Research Funding; Umoja: Consultancy; Genentech: Consultancy, Research Funding; Bristol Myers Squibb: Consultancy, Research Funding; Novartis: Consultancy, Research Funding; Kite, a Gilead Company: Consultancy, Research Funding; MorphoSys: Consultancy, Research Funding; Iksuda Therapeutics: Consultancy; ADC Therapeutics: Consultancy, Research Funding; AstraZeneca: Consultancy, Research Funding; Morphosys: Research Funding; 47 Inc: Research Funding.